Parallel filtering and smoothing in an LG-SSM#
This notebook shows how can reduce the cost of inference from linear to logarithmic in time, if we have a GPU device. Rather than running a Kalman filter/smoother sequentially in time, you can use a parallel (associative) scan to compute the filtering and smoothing distributions using a parallel algorithm that requires \(O(T)\) parallel processors, but can run in only \(O(\log T)\) time. This is the key innovation behind modern “deep” state space models like S5 (Smith et al., 2023) and Mamba (Gu and Dao, 2024). The core algorithm is nicely described by Särkkä and Garćıa-Ferńandez (2020).
This notebook is adapted from an example notebook by Adrien Correnflos. Some small changes have been made so it works with dynamax.
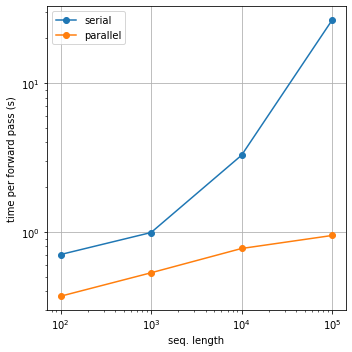
If you have a GPU, you should be able to get a speedup curve like this:
Similar algorithms exist for HMMs (Hassan et al., 2021), which we have implemented in dynamax.hidden_markov_model.parallel_inference
References#
Särkkä, Simo, and Ángel F. García-Fernández. “Temporal parallelization of Bayesian smoothers.” IEEE Transactions on Automatic Control 66.1 (2020): 299-306.
Smith, Jimmy TH, Andrew Warrington, and Scott Linderman. “Simplified State Space Layers for Sequence Modeling.” The Eleventh International Conference on Learning Representations.
Gu, Albert, and Tri Dao. “Mamba: Linear-time sequence modeling with selective state spaces.” arXiv preprint arXiv:2312.00752 (2023).
Hassan, Syeda Sakira, Simo Särkkä, and Ángel F. García-Fernández. “Temporal parallelization of inference in hidden Markov models.” IEEE Transactions on Signal Processing 69 (2021): 4875-4887.
Setup#
import jax
from jax import numpy as jnp
from jax import random as jr
from jax import block_until_ready
from matplotlib import pyplot as plt
import time
from dynamax.linear_gaussian_ssm import lgssm_smoother, parallel_lgssm_smoother
from dynamax.linear_gaussian_ssm import LinearGaussianSSM
print(jax.devices())
print(jax.devices()[0].platform)
if jax.devices()[0].platform == 'cpu':
cpu_mode = True
else:
cpu_mode = False
[CpuDevice(id=0)]
cpu
Model#
The model is a simple tracking model (see Example 3.6 in Bayesian Filtering and Smoothing (S. Särkkä, 2013).
dt = 0.1
F = jnp.eye(4) + dt * jnp.eye(4, k=2)
Q = 1. * jnp.kron(jnp.array([[dt**3/3, dt**2/2],
[dt**2/2, dt]]),
jnp.eye(2))
H = jnp.eye(2, 4)
R = 0.5 ** 2 * jnp.eye(2)
μ0 = jnp.array([0.,0.,1.,-1.])
Σ0 = jnp.eye(4)
latent_dim = 4
observation_dim = 2
input_dim = 0
lgssm = LinearGaussianSSM(latent_dim, observation_dim)
params, _ = lgssm.initialize(
initial_mean=μ0,
initial_covariance= Σ0,
dynamics_weights=F,
dynamics_covariance=Q,
emission_weights=H,
emission_covariance=R)
Test parallel inference on a single sequence#
num_timesteps = 100
key = jr.PRNGKey(0)
key, subkey = jr.split(key)
z,emissions = lgssm.sample(params, subkey, num_timesteps)
ssm_posterior = lgssm_smoother(params, emissions)
parallel_posterior = parallel_lgssm_smoother(params, emissions)
print(parallel_posterior.filtered_means.shape)
print(parallel_posterior.filtered_covariances.shape)
(100, 4)
(100, 4, 4)
assert jnp.allclose(parallel_posterior.filtered_means, ssm_posterior.filtered_means, atol=1e-3)
assert jnp.allclose(parallel_posterior.filtered_covariances, ssm_posterior.filtered_covariances, atol=1e-3)
assert jnp.allclose(parallel_posterior.smoothed_means, ssm_posterior.smoothed_means, atol=1e-3)
assert jnp.allclose(parallel_posterior.smoothed_covariances, ssm_posterior.smoothed_covariances, atol=1e-3)
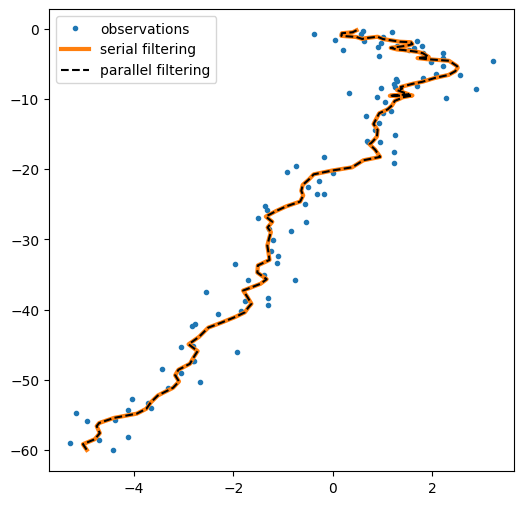
plt.figure(figsize=(6,6))
plt.plot(*emissions.T,'.', label="observations")
plt.plot(*ssm_posterior.filtered_means[:,:2].T, label="serial filtering", lw=3)
plt.plot(*parallel_posterior.filtered_means[:,:2].T, "--k", label="parallel filtering");
plt.legend();
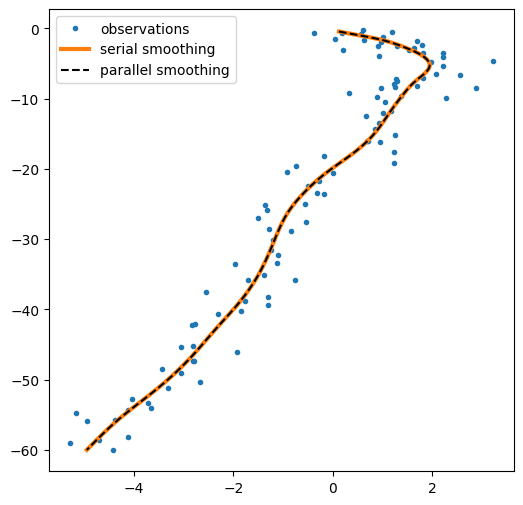
plt.figure(figsize=(6,6))
plt.plot(*emissions.T,'.', label="observations")
plt.plot(*ssm_posterior.smoothed_means[:,:2].T, lw=3,label="serial smoothing")
plt.plot(*parallel_posterior.smoothed_means[:,:2].T, "--k", label="parallel smoothing")
plt.legend();
Timing comparison#
key = jr.PRNGKey(0)
if cpu_mode:
Ts = [100, 200, 500]
num_repeats = 1
else:
Ts = [100, 1_000, 10_000, 100_000]
num_repeats = 3
serial_smoothing_durations = []
parallel_smoothing_durations = []
compiled = False
for T in Ts:
key, subkey = jr.split(key)
z,emissions = lgssm.sample(params, subkey, T)
if not compiled:
ssm_posterior = block_until_ready(lgssm_smoother(params, emissions))
parallel_posterior = block_until_ready(parallel_lgssm_smoother(params, emissions))
start = time.time()
for _ in range(num_repeats):
ssm_posterior = block_until_ready(lgssm_smoother(params, emissions))
end = time.time()
mean_time = (end-start)/num_repeats
serial_smoothing_durations.append(mean_time)
print(f"Num timesteps={T}, \t time serial = {mean_time}")
start = time.time()
for _ in range(num_repeats):
parallel_posterior = block_until_ready(parallel_lgssm_smoother(params, emissions))
end = time.time()
mean_time = (end-start)/num_repeats
parallel_smoothing_durations.append(mean_time)
print(f"Num timesteps={T}, \t time parallel = {mean_time}")
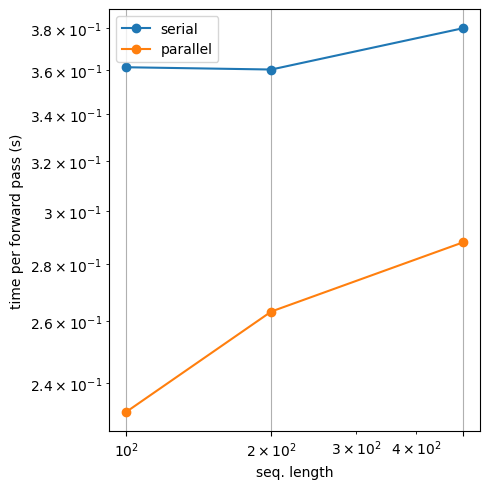
Num timesteps=100, time serial = 0.3613753318786621
Num timesteps=100, time parallel = 0.23122191429138184
Num timesteps=200, time serial = 0.36031079292297363
Num timesteps=200, time parallel = 0.26334381103515625
Num timesteps=500, time serial = 0.3801305294036865
Num timesteps=500, time parallel = 0.28804779052734375
plt.figure(figsize=(5, 5))
plt.loglog(Ts, serial_smoothing_durations, '-o', label='serial')
plt.loglog(Ts, parallel_smoothing_durations, '-o', label='parallel')
plt.xticks(Ts)
plt.xlabel("seq. length")
plt.ylabel("time per forward pass (s)")
plt.grid(True)
plt.legend()
plt.tight_layout()